The Circuitry in Our Cells
As you sit quietly, trillions of cells in your body are performing a frenetic dance of biochemical computation that makes your existence possible.
Consider this: You were once a single cell — a fertilized egg. This single cell was equipped with a genetic program capable of assembling atomically-precise molecular machines, replicating and distributing copies of its genetic program through cell division, and self-organizing multicellular structures into a human shape with specialized cell types, tissues, and organs.
And now here you are, reading this: Your eyes scanning these words while your brain interprets them. You built yourself from scratch.

Biology computes with genetic circuits
The remarkable ability of biology to create patterns, perform specialized tasks, and adapt to changing environments is made possible with genetic circuits — networks of interacting genes that perform computation.
Genetic circuits appear literally everywhere in nature. In a lone bacterium as it “tumbles and runs” toward food. In a California redwood as it constructs itself into the sky. And in your immune system as it wards off cancer and infection. In fact, every single thing that civilization sources from biology — food, materials, drugs — was built by nature using genetic circuits to exert fine spatiotemporal control over biochemistry.
Yet despite their ubiquity in nature, genetic circuits are not harnessed in most biotechnology today. Instead, the state-of-the-art is constant overproduction of a few genes, whether they be enzymes, pesticides, or peptides.
Future biotechnologies will seem like science fiction: Intelligent therapeutics programmed to sense disease in the human body and trigger a therapeutic response. Living materials that can heal and react to their surroundings. Smart plants that can modify their physiology to withstand extreme cold or drought. To make these biotechnologies a reality, we need to be able to engineer genetic circuits.
From discovery to design
Natural genetic circuits have been studied for more than half a century. In 1961, the French scientists François Jacob and Jacques Monod published a landmark paper describing the genetic circuit in E. coli that senses and eats lactose [1]. Their description of how the appropriate metabolic genes are regulated (known as the lac operon model) was the first of its kind.
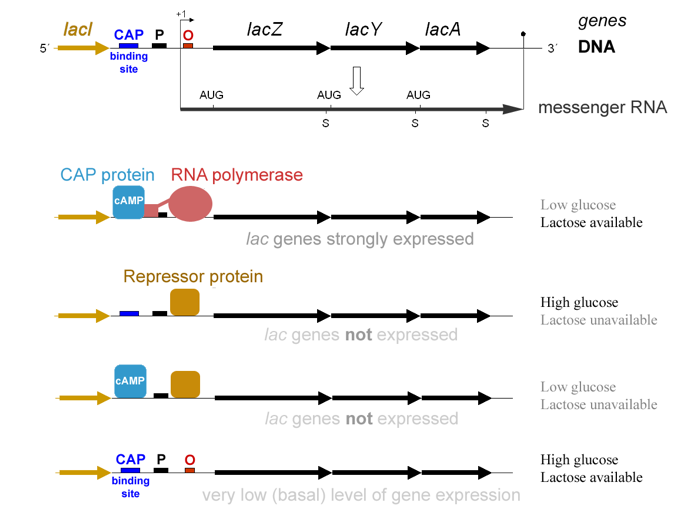
A few months later, they predicted that similar regulatory processes could explain cell differentiation in multicellular organisms, like humans. Without mincing words, they wrote, “Moreover, it is obvious from the analysis of these mechanisms that their known elements could be connected into a wide variety of ‘circuits’, endowed with any desired degree of stability” [2]. For their work, they were awarded the Nobel Prize for Physiology or Medicine in 1965, along with André Lwoff.
In the years since that seminal discovery, scientists have further illuminated the myriad ways biological systems achieve behavior — from everyday tasks to exceptional feats. Indeed, entire books have been written on natural genetic circuits. (Check out the classic, “A Genetic Switch” by Mark Ptashne [3], which describes how the bacterial virus lambda phage regulates its life cycle.) The panoply of molecular mechanisms that power biological computation is vast and diverse, and reverse engineering natural genetic circuits is a field of intense ongoing research.
Armed with insights from nature, biological engineers began to design synthetic genetic circuits from the ground up. Back-to-back publications in Nature in 2000 are considered by many to be the first examples in the field (a genetic oscillator [4] and toggle switch [5]).
Over the past two decades, the ability to engineer increasingly complex and precise genetic circuits has advanced rapidly. Progress has resulted from several factors: thousands of sequenced genomes (and metagenomes) from which to “mine” useful genes, faster and cheaper DNA synthesis and sequencing, an improved understanding of cell biophysics to enable simulation, the ability to make targeted genomic modifications using CRISPR, and last but not least, years of compounded genetic engineering experience distilled into guiding design principles.
We are truly in the early days of a golden era for engineering biology.
Yet despite our progress so far, genetic circuit design has often been characterized by a manual and failure-prone process. Engineers often spend years creating a functional design through trial-and-error.
Automating genetic circuit design
How might this process of genetic circuit design be systematized and made more reliable? The semiconductor industry has completely transformed society, and its evolution offers a case study in transitioning from artisanal to automated.

Early on, electronics engineers would painstakingly design and lay out circuit diagrams by hand. Then, the 1970s brought with it the first taste of automation: “place and route” techniques developed to position all of the electronic components and wires.
In the 1980s, the advent of electronic design automation (EDA) enabled programming languages that could be compiled down to patterns in silicon. One of the early publications describing this capability, “Introduction to VLSI Systems” by Carver Mead and Lynn Conway, is a holy text of EDA [6]. This breakthrough drove rapid increases in electronic chip complexity, and EDA became an entire industry in itself.
Today, chip designers use sophisticated EDA software that automates the entire workflow (design, simulation, and manufacturing). Software was truly brought to bear on electronic circuit design, and was one of the key enablers of Moore’s law.
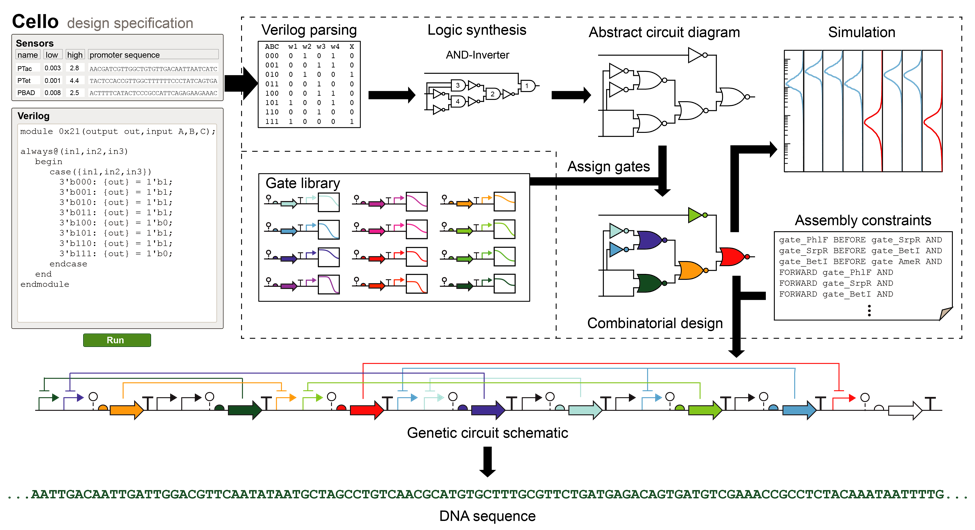
Drawing inspiration from this evolution, we built a genetic circuit design automation platform, Cello (short for “Cell Logic”) [7]. We even used a common electronic hardware description language (Verilog) from electronics design to write our circuit specifications.
By combining concepts from digital logic synthesis, cell biophysics, and synthetic biology, we were able to build genetic circuits with up to 10 interacting genes. That’s state-of-the-art for an engineered cell behavior in 2017, but it still pales in comparison to nature. For reference, the E. coli genome employs roughly 300 genes called transcription factors to control metabolism, survival, and replication. Human cells have about an order of magnitude more. (While this may seem paltry compared to the billions of transistors in a modern CPU, it’s an apple and oranges comparison. The point isn’t to compete with silicon — the point is to program biology with new functions.)
A tremendous amount of engineering lies ahead before we achieve genome-scale design with comparable complexity, elegance, and subtlety to what nature has evolved. We’re working on that. On the other hand, we have reached a point where genetic circuit engineering is reliable enough that we can program cell functions for previously impossible biotechnologies.
Introducing Asimov
In the same way that electronic circuits have become ubiquitous in the world — from cars to mobile phones to smart refrigerators — the same will become true for engineered genetic circuits. They will begin to appear in many aspects of daily life, from therapeutics to agriculture to consumer goods.
To help lay the foundation, I’m proud to announce the launch of Asimov with my co-founders Chris Voigt, Doug Densmore, and Raja Srinivas. Building upon our initial work on Cello, we’re developing a platform for professional genetic circuit design. We strive for Asimov to be the go-to resource for designing biological computation as biotechnology steadily becomes a fully-fledged engineering discipline.
I personally hope that this technology one day improves our ability to cure disease, empowers clean and sustainable manufacturing, and helps nourish a growing global population.
I look forward to keeping you updated on our progress.

.png)



